Using DrugBank Structured Pharmacology Data
Introduction
Pharmacology data captures a drug's journey through the body by measuring how it's absorbed, distributed, metabolized, and eliminated. To date, many studies with pharmacology data and clear experimental conditions have been published, but this information is not available in a machine-readable format. For it to be used in pharmacokinetic modelling or other applications, data must be retrieved and structured through curation efforts. We have extracted and structured pharmacology data from our free-text data so that it is easier to process and analyze.
This structured data is available in a series of tables called the structured_pharmacology_ tables. This guide will help you understand the value of this data and how it can be used in your research.
How Is This Data Organized?
| Parameter | Description | Table Name(s) |
|---|---|---|
| Area under the curve (AUC) | Measures the total systemic exposure to a drug over time. | structured_pharmacology_aucs |
| Bioavailability | Measures the rate and extent to which a drug reaches systemic circulation, typically as a percentage. | structured_pharmacology_bioavailabilities |
| Clearance | Measures the rate at which a drug is removed from the body. | structured_pharmacology_clearances |
| Maximum concentration (Cmax) | Measures the peak serum concentration reached by a drug | structured_pharmacology_cmaxes |
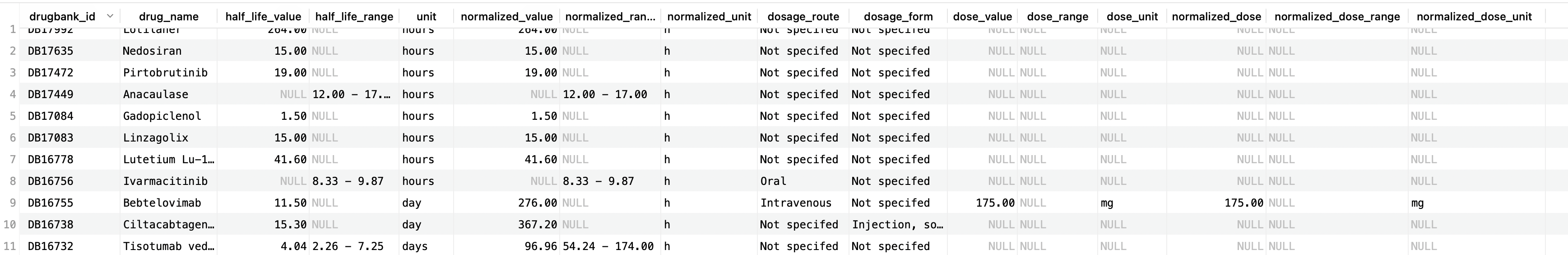
| Half-life | Measures the time for a drug's concentration to be reduced by half. | structured_pharmacology_half_lives |
Each value can be matched to the drug it corresponds to using the drug identifier (drug_id). Values are provided in both their original reported units and as normalized values. When reported by a study, we also include the dosages used to determine these pharmacology parameters and subject characteristics, such as age and sex.
For more information about the data covered for each parameter, as well as the columns included in each of these tables, go to our CSV format documentation that covers Structured Pharmacologies.
How Can This Be Used?
The structured format of this data allows you to perform queries and analysis without first needing to extract it in a machine-readable format. This enables you to:
- Query and analyze specific values, such as a drug's half-life or clearance rate
- Compare pharmacokinetic properties across different drugs, dosage forms, or patient populations
- Integrate this data more easily into your workflows and applications and match it to values obtained from different sources
This is an example of a query that pulls half-life data:
SELECT d.drugbank_id, d.name AS drug_name, TRUNCATE(sphl.value,2) AS half_life_value, CONCAT(TRUNCATE(sphl.min,2), ' - ', TRUNCATE(sphl.max,2)) AS half_life_range, sphl.unit, TRUNCATE(sphl.normalized_value,2) AS normalized_value, CONCAT(TRUNCATE(sphl.normalized_min,2), ' - ', TRUNCATE(sphl.normalized_max,2)) AS normalized_range, sphl.normalized_unit, COALESCE(dr.name, 'Not specified') AS dosage_route, COALESCE(df.name, 'Not specified') AS dosage_form, TRUNCATE(sphl.dose_value,2) AS dose_value, CONCAT(TRUNCATE(sphl.dose_min,2), ' - ', TRUNCATE(sphl.dose_max,2)) AS dose_range, sphl.dose_unit, TRUNCATE(sphl.normalized_dose_value,2) AS normalized_dose, CONCAT(TRUNCATE(sphl.normalized_dose_min,2), ' - ', TRUNCATE(sphl.normalized_dose_max,2)) AS normalized_dose_range, sphl.normalized_dose_unit FROM drugs d JOIN structured_pharmacology_half_lives sphl ON d.id = sphl.drug_id LEFT JOIN dosage_routes dr ON sphl.dosage_route_id = dr.id LEFT JOIN dosage_forms df ON sphl.dosage_form_id = df.id GROUP BY sphl.id;Copy to clipboard
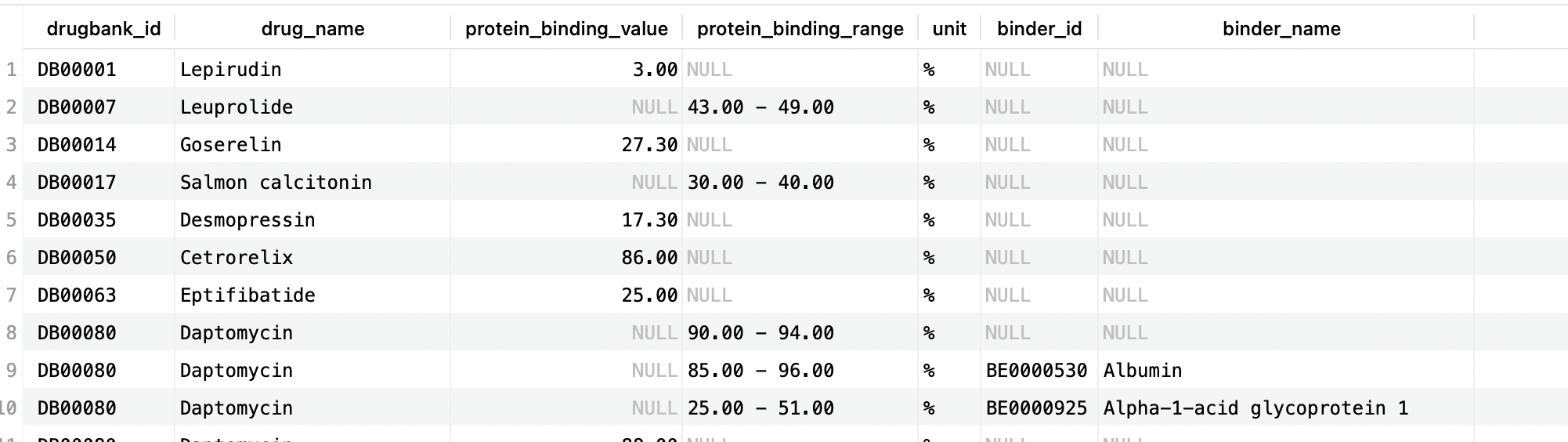
Queries for other parameters such as AUC, Cmax, and clearance will follow a similar structure since each structured_pharmacology_ table will include all relevant information. For protein bindings, both structured_pharmacology_protein_bindings and structured_pharmacology_protein_bindings_bio_entities are needed:
SELECT d.drugbank_id, d.name AS drug_name, TRUNCATE(pb.normalized_value,2) AS protein_binding_value, CONCAT(TRUNCATE(pb.normalized_min,2), ' - ',TRUNCATE(pb.normalized_max,2)) AS protein_binding_range, pb.unit, be.biodb_id AS binder_id, be.name AS binder_name -- binder_name provided if value specific to a single protein FROM structured_pharmacology_protein_bindings pb LEFT JOIN drugs d ON pb.drug_id = d.id LEFT JOIN structured_pharmacology_protein_bindings_bio_entities sbe ON pb.id = sbe.structured_pharmacology_protein_binding_id LEFT JOIN bio_entities be ON sbe.bio_entity_id = be.biodb_id GROUP BY pb.id, be.biodb_id;Copy to clipboard
Why Does It Matter
Pharmacology data is used in a variety of applications, including drug development, clinical trial design, and personalized medicine. With our structured pharmacology dataset, you can integrate these parameters directly into your models without extensive data preparation, boosting both your speed and accuracy. This streamlined approach means you will spend less time cleaning data and more time on high-impact analysis and discovery. All parameters of our structured pharmacology dataset are referenced, making it easier to go back to the original source of information. With data readily available in a structured format, you can accelerate the entire research pipeline, from initial hypothesis to new drug insights.
Conclusion
With our structured pharmacokinetic data, you get a resource that’s accurate, accessible, and ready to use. Because the data comes in normalized units and with detailed context, you can run deeper analyses and draw more reliable insights into drug behavior. This helps you streamline research and development, integrate high-quality data into your workflows, and ultimately accelerate innovation.